For UK and Ireland healthcare professionals only.
How-to guide for healthcare professionals: understanding genomic reports
This educational genomic report reader tool microsite has been fully initiated and funded by Boehringer Ingelheim. It was co-developed in collaboration with Springer Health+, who were commissioned to create the tool alongside expert healthcare professionals who received honoraria for their contributions. Boehringer Ingelheim provided strategic guidance throughout the development process and reviewed the content to ensure compliance with the ABPI Code of Practice.
Genomics report overview
Watch this 3 minute video, featuring Dr Adam Januszewski and Dr Igor Gomez-Randulfe, to find out more about the importance of understanding the nuances of genomic reports.
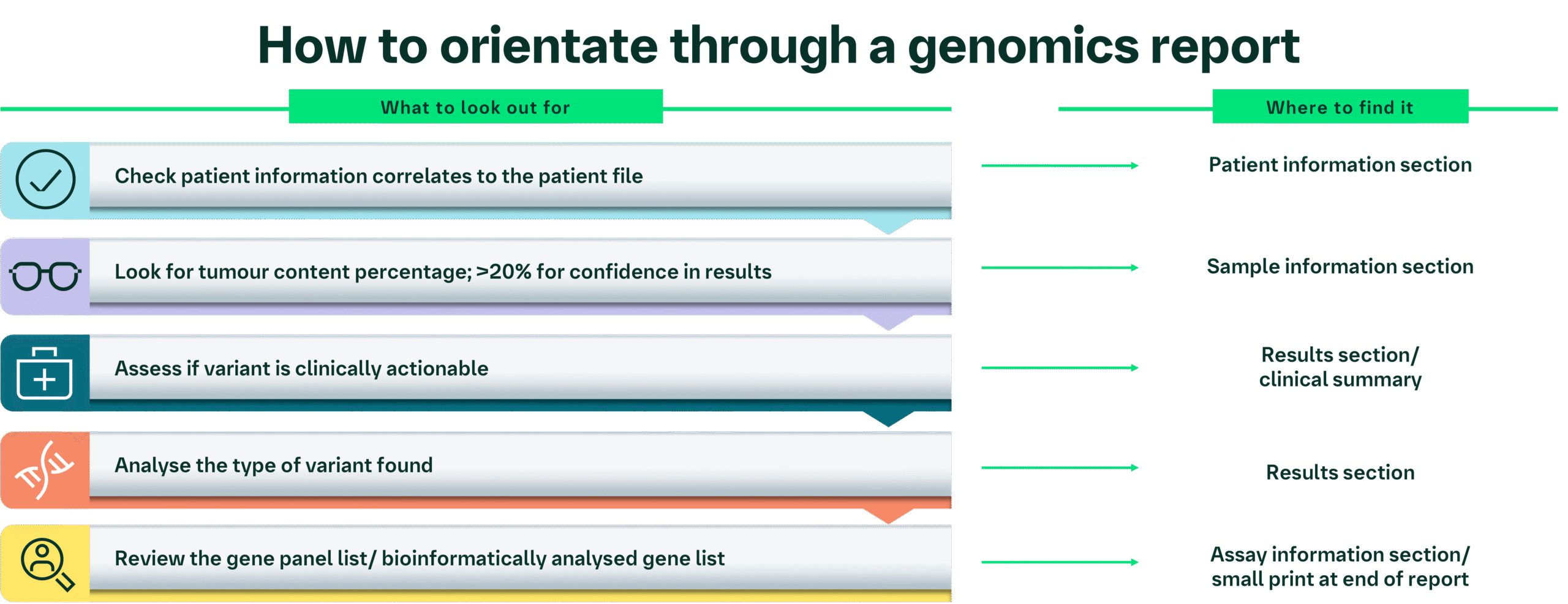
Genomic reports can vary significantly in the way that they present information; however, presented below is a flow chart to help navigate through the content you would expect to find in a report.
Orientation: key elements of a genomic report
Name: Jon Smith
Date of birth: 04/01/1963
Gender: Male
NHS number: 1234567890
Local hospital number: 123456X
Report Date: 16/07/2025
Note: The information presented here is for a fictional patient and is intended only for educational purposes; it does not intend to reflect an individual patient.
This section includes patient identification information and is usually found at the top of a report. You need to double check this information correlates with your files to ensure that this is the correct report for your patient.
There are multiple reports that can be issued for a patient (for example, for DNA analysis and RNA analysis), which may be in a sequential order. All the reports will all need to be reviewed; however, it is important to ensure that the date of the report is taken into account, so any additional reports that may have been created are not missed. It is also important to read the whole document.
Biopsy date: 06/07/2025
Biopsy number: Slides received from biopsy 123
Tumour type: Non-small cell lung cancer
Neoplastic cell content was estimated to be 40%
Details about the sample are presented in this section. It is important to note that the type of information varies between different reports; however, a key point to focus on is checking that the correct tumour type panel has been carried out (e.g., a lung cancer panel for a non-small cell lung cancer [NSCLC] sample).
This section shows the biopsy date, which is the date the sample was collected. This is often different from the analysis/ report date, as it may take some time to process the sample histologically. Moreover, the laboratory may run multiple samples in one assay, so they may wait for enough samples to be collected to make the process more efficient.
In some instances, there may be a pathology number listed on the report that relates specifically to the sample/biopsy, which is also affiliated with the patient information.
The report includes the type of tumour analysed. Next-generation sequencing (NGS) is performed for different types of cancers such as lung, breast, colorectal, etc., to see if a patient has a specific mutation for which there may be targetable treatment, if a different treatment regimen is suitable, or if they are suitable for entry into a clinical trial.
The tumour content/percentage of tumour cells present within the sample is usually provided by the pathologist before the sample is run on an NGS panel. This is an important part of the report.
Watch this 45 second video to find out about the importance of tumour content.
If the tumour content is <20%, the GLH may refuse to run the analysis1‒3, because the NGS analysis may fail to detect mutations due to insufficient DNA content. Of note, Genomics England classifies <40% as “low” tumour content4.
If the sample does undergo NGS analysis, then caution should be taken when determining if this is a true negative result, as there may not be enough DNA present within the sample to detect a mutation.
In some instances, if the tumour content is <10–20%, repeat biopsy and genomic testing may be required for confirmation of the results. However, this may not be possible if the tumour is in an inaccessible location, or if the patient has already started treatment.
Other salvage techniques, such as polymerase chain reaction (PCR) testing, may be an option if re-biopsy is not possible as this process amplifies the DNA present within the sample; however, this may not be available in all Trusts.
If the tumour content is high, then you can be more confident in the validity of the NGS results.
- NHS East Genomics. Specimen requirements – solid cancer. Accessed December 2025. https://www.eastgenomics.nhs.uk/for-healthcare-professionals/genomic-tests/referral-forms-index/cancer-solid-tumour-tests-non-wgs/specimen-requirements-solid-cancer/.
- NHS North East and Yorkshire GLH, solid cancer genomics referral form. Accessed December 2025. https://ney-genomics.org.uk/wp-content/uploads/2023/09/Solid_cancer_genomics_referral_form_Aug23.pdf.
- NHS South East Genomics. Solid tumour other; order or find a test. Accessed December 2025. https://southeastgenomics.nhs.uk/tool/solid-tumour-other-i-e-specific-histology-not-listed-elsewhere-in-the-test-directory/.
- Genomics England. GMS Interpretation Portal. Cancer referral overview page. Accessed December 2025. https://ip-portal-help.genomicsengland.co.uk/1.1/GMS-Interpretation-Portal/gms_cancer_referral_overview/#:~:text=Tumour%20content%20is%20displayed%20as,the%20point%20of%20test%20order
Analysis/report date: 16/07/2025
Technical information: Analysis was carried out using the Illumina TSO500 NGS gene panel.
Disclaimer: The Illumina NGS panel presented here is one example vendor; other reports may use different vendors
In this example report, the gene panel sequenced 523 genes, with the aim of detecting small nucleotide variants, copy number variants, and gene fusions. Of these, 50 genes were analysed through the South-West GLH in-house bioinformatic somatic pipeline (for NSCLC).
Specific coverage of any particular panel will vary from Centre to Centre, but any given panel will include the National Genomic Test Directory targets at a minimum.1
The analysis or report date will tell you when the bioinformatic analysis was undertaken. This may be different from the biopsy date.
The type of analysis conducted may be found within the technical information section or notes section in small print.
NGS is a method/technology for analysing DNA or RNA sequences, in order to study genetic variation. It is a highspeed process that can sequence millions of fragments of sequences at the same time, with high accuracy.2
DNA NGS is used to sequence part of the genome and can provide insights into inherited gene mutations, single-nucleotide polymorphisms and small insertions or deletions. Whereas RNA NGS provides insights into the transcriptome, gene expression levels, and gene fusions/recombinants.2
Other genomic analyses may be carried out, depending on disease features and stage of disease. For example, circulating tumour DNA (ctDNA) assays, which look at the DNA content that may be released by a tumour into the blood, or PCR testing (if the tumour content is <10–20%). These types of analysis will have their own reports.
Watch this 1-minute video to find out about the importance of the gene panel and how it may vary.
The gene panel may be specified at the beginning of the report i.e., as “Non-small cell lung cancer gene panel was requested”, then explained in more detail in the technical information in small print or at the end of the report on a separate page.
The number of genes included in the panel may vary depending on which laboratory or hub is carrying out the testing, but can be up to ~500 genes, ~200 genes, or ~50 genes (as they tend to cover important genes for different types of cancers).
Bioinformatic analysis is then only carried out on a smaller subset of genes (usually about 8–16 genes, possibly up to ~50), which are specific for the type of cancer the patient has.
It is stipulated that anything in the National Genomic Test Directory that is a gene associated with the cancer type in question (e.g., NSCLC) has to be reported; additional genes included in the gene panel vary across the GLHs.
Genes that are bioinformatically analysed after DNA NGS for NSCLC include:1
- EGFR
- BRAF
- KRAS
- MET
- ERBB2
There are also genes that are analysed using RNA NGS, such as ALK, RET, and NTRK1/2/3. These may feature in an RNA report.
Some reports will then only list those genes where a mutation was found that is clinically actionable (has a targeted treatment available) in the results section.
- NHS England. National Genomic Test Directory for Cancer. Published 9 July 2025. Accessed December 2025. https://www.england.nhs.uk/publication/national-genomic-test-directories/
- Satam H, Joshi K, Mangrolia U, et al. Next-generation sequencing technology: current trends and advancements. Biology (Basel). 2023;12(7):997.
Gene: EGFR
Variant(s): EGFR c.2573T>G p.(Leu858Arg)
The report will show the genes that were analysed bioinformatically and the mutation found, if any. This information may be clustered together in a table or as a paragraph of text.
In this example you can see the patient has a mutation in the EGFR gene. Other examples of commonly reported genes include KRAS, BRAF, MET, etc.
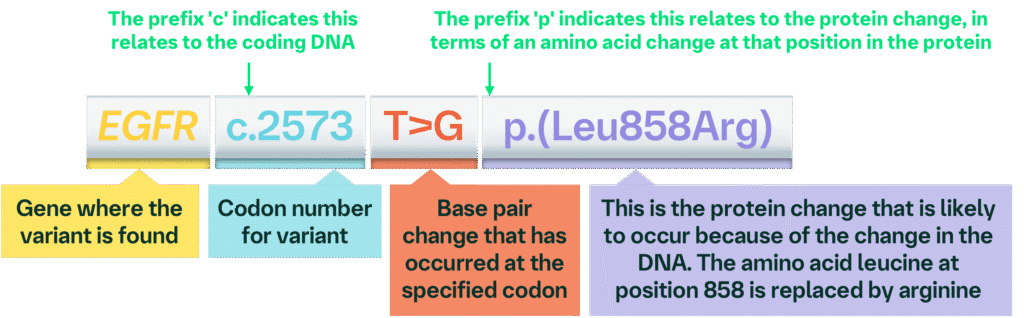
Taken together, this variant information tells you that there has been a base pair change to the EGFR gene at codon 2573, where a thymine (T) is replaced by guanine (G), which results in an amino acid change in the protein sequence at position 858 from leucine to arginine.
This example may also appear using a single letter amino acid code: EGFR c.2573 T>G p.L858R (see below for more information about amino acid codes).
Other examples of different types of annotations include:
EGFR p.(Asp770_Asn771insGly)
This tells you that, for the EGFR gene, there is a genetic variation that results in an amino acid insertion of glycine between the aspartic acid at position 770 and asparagine at position 771.
EGFR p.E746_A750del
This tells you that, for the EGFR gene, the genetic variation results in a deletion between the glutamic acid at position 746 and alanine at position 750.
Watch this 1-minute video to find out how the amino acid code used may vary in a genomics report.
It is important to note that the amino acid change may be denoted as a three-letter code or a one-letter code, depending on how the individual laboratory reports their results. Therefore, it is essential to know the difference, and that the first letter of the three-letter code does not always correlate to the one-letter code.
Amino acid coding:1
| Amino acid | Three-letter code | One-letter code |
|---|---|---|
| Alanine | Ala | A |
| Arginine | Arg | R |
| Asparagine | Asn | N |
| Aspartic acid | Asp | D |
| Cysteine | Cys | C |
| Glutamic acid | Glu | E |
| Glutamine | Gln | Q |
| Glycine | Gly | G |
| Histidine | His | H |
| Isoleucine | Ile | I |
| Leucine | Leu | L |
| Lysine | Lys | K |
| Methionine | Met | M |
| Phenylalanine | Phe | F |
| Proline | Pro | P |
| Serine | Ser | S |
| Threonine | Thr | T |
| Tryptophan | Trp | W |
| Tyrosine | Tyr | Y |
| Valine | Val | V |
- Pure and Applied Chemistry. A one-letter notation for amino acid sequences (definitive rules). 1972;31(4):639‒646.
Watch this animation to find out about the different types of genomic aberrations detected by NGS.1-3
- National Human Genome Research. Talking glossary of genomic and genetic terms. Accessed December 2025. https://www.genome.gov/genetics-glossary
- National Library of Medicine. The NCBI handbook. 2nd Edition. 2013. Accessed December 2025. https://www.ncbi.nlm.nih.gov/sites/books/NBK470040/
- Genomics Education Programme. Genomics Glossary. Accessed December 2025. https://www.genomicseducation.hee.nhs.uk/glossary/
Variant allele frequency (VAF [%]): 30%
Alleles are alternative forms of a gene at the same position on a chromosome. The VAF is a measure of the fraction of alleles that have the genomic variant relative to the total number of alleles within a genomic locus. It may represent a surrogate for heterogeneity of a tumour.1
A high VAF suggests that a high proportion of tumour cells harbour the genetic variation and therefore there is a dominant clone. A high VAF may also be reflective of a driver mutation (one that directly contributes to the development and progression of cancer), as the mutation provides a selective advantage over other sub-clones within the tumour and would therefore become more prevalent over time (as the tumour grows).1
Conversely, a low VAF, alongside high tumour content, suggests heterogeneity of the tumour, thus the targeted therapy may be less effective because of the presence of other subclones within the tumour.1
It is important to note that the VAF and tumour content are two separate metrics. The former indicates the proportion of variant DNA in the sample, while the latter reflects how many tumour cells are present in the sample. Both should be interpreted in their own right, side by side. This allows for interpretation in terms of confidence levels in the findings.
VAF can also help infer if the mutation is germline or somatic. If the VAF is high (around 50% or higher) it is likely to be a germline mutation and therefore inherited.2 This is because either one or both parents passed on the mutation, resulting in heterozygous or homozygous inheritance, respectively.2
Watch this 30-second video about the difference between somatic and germline mutations.
- Bielo LB, Trapani D, Repetto M, et al. Variant allele frequency: a decision-making tool in precision oncology? Trends Cancer. 2023;9(12):1058–1068.
- Kraft IL, and Godley LA. Identifying potential germline variants from sequencing hematopoietic malignancies. Hematology Am Soc Hematol Educ Program. 2020;2020(1):219–227.
- NHS England. National Genomics Education programme. Constitutional (germline) vs somatic (tumour) variants. Accessed December 2025. https://www.genomicseducation.hee.nhs.uk/genotes/knowledge-hub/constitutional-germline-vs-somatic-tumour-variants/
Clinical summary: Molecular analysis detected the EGFR activating variant in exon 21 of the EGFR gene in a high proportion of cells in the tumour sample. This indicates the patient may benefit from treatment with EGFR tyrosine kinase inhibitor (TKI) targeted therapy.
It is important to discuss the genomic report with the pathologists in your multidisciplinary team (MDT), for example, to address false negatives vs true negatives. This meeting is an opportunity to delve into some of the common challenges and pitfalls faced, for instance where there are variants identified with DNA analysis that are not found with RNA analysis and to provide support with cases.
Watch this 1-minute video to find out about the importance of mutations with clinical significance.
You may find the clinical summary at the top of the report in a boxed section or in bold. Specifics for this section are likely to vary from Centre to Centre, as well as between different variants. However, this information will show you the mutations identified by NGS that have a clinical significance or those with potential clinical significance. These may be listed in a tiered system on some reports. Those that are considered tier III onwards are unlikely to be included in a genomics report.
| Tier | Clinical significance | ESCAT description1 |
|---|---|---|
| I | Strong | Alteration-drug match is associated with improved outcome in clinical trials |
| II | Potential | Alteration-drug match is associated with antitumour activity, but magnitude of benefit is unknown |
| III | Unknown | Alteration-drug match is suspected to improve outcome based on clinical trial data in other tumour type(s) or with similar molecular alteration |
| IV | Benign/likely benign | Preclinical evidence of actionability |
| V | None | Alteration-drug match is associated with objective response, but without clinically meaningful benefit |
| X | None | Lack of evidence for actionability |
ESCAT: ESMO scale of clinical actionability for molecular targets.
- Mateo J, Chakravarty D, Dienstmann R, et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann Oncol. 2018;29(9):1895–1902.
Some reports may list specific treatments that are suitable for the specific mutation(s) detected, other reports may just state the drug class that may be appropriate (as seen here, e.g., tyrosine kinase inhibitors).
Sometimes references supporting the clinical evidence for the suggested treatment options are also listed on the report; however, caution should be taken to make sure this information is up to date.
Gene panel analysed for NSCLC: EGFR, MET, BRAF, KRAS, ERBB2, PIK3CA.
The gene panel list may be found in the technical information in small print, or at the end of the report on a separate page. It shows all the genes that were included in the NGS run (but not all will be bioinformatically analysed). RNA reports may include other genes such as ROS1, RET, ALK, NTRK1/2/3, NRAS, AKT1, PTEN, ARID1A, ARID2, ATM, BRCA1, BRCA2, BRIP1, CCND1, CCND3, CCNE1, CDK12, CDK4, CDKN2A, ERBB3, ERBB4, FGFR1, FGFR2, FGFR3, KEAP1, MAP2K1 (MEK1), MDM2, NF1, NFE2L2, NRG1, PALB2, RAD51C, RAD51D, RB1, RBM10, RICTOR, RIT1, SETD2, SMARCA4, STK11, TP53, TSC1/2, U2AF1. The number of genes listed here may vary, as described in the ‘assay information’ section.
Please take a moment to test your current knowledge. Select one answer for each question.